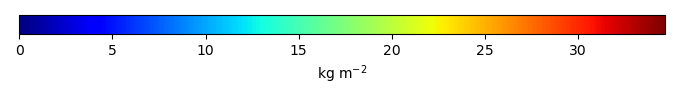
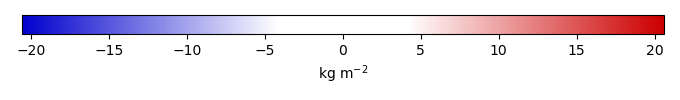
Mean State
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 30.1 | ||||||||||
| bcc-csm1-1 | [-] | 476. | 25.2 | 443. | 30.1 | 0.000275 | -0.292 | 0.568 | 0.368 | 0.468 | ||
| bcc-csm1-1-m | [-] | 385. | 21.4 | 363. | 29.9 | 0.201 | -0.843 | 0.546 | 0.368 | 0.457 | ||
| BNU-ESM | [-] | 917. | 69.1 | 831. | 30.1 | 2.95e-05 | 4.75 | 0.427 | 0.518 | 0.472 | ||
| CanESM2 | [-] | 529. | 21.3 | 495. | 30.1 | -0.948 | 0.566 | 0.730 | 0.648 | |||
| CESM1-BGC | [-] | 529. | 18.8 | 509. | 29.9 | 0.176 | -1.03 | 0.601 | 0.659 | 0.630 | ||
| GFDL-ESM2G | [-] | 655. | 28.9 | 618. | 30.1 | 0.191 | 0.534 | 0.620 | 0.577 | |||
| HadGEM2-CC | [-] | 439. | 19.3 | 418. | 29.8 | 0.282 | -0.790 | 0.606 | 0.669 | 0.638 | ||
| HadGEM2-ES | [-] | 453. | 19.1 | 431. | 29.8 | 0.282 | -0.818 | 0.602 | 0.664 | 0.633 | ||
| inmcm4 | [-] | 603. | 63.4 | 542. | 28.7 | 1.37 | 3.99 | 0.462 | 0.824 | 0.643 | ||
| IPSL-CM5A-LR | [-] | 622. | 44.3 | 567. | 30.1 | 0.0202 | 2.24 | 0.542 | 0.780 | 0.661 | ||
| IPSL-CM5A-MR | [-] | 633. | 43.8 | 583. | 30.1 | 0.0426 | 2.05 | 0.551 | 0.775 | 0.663 | ||
| MeanCMIP5 | [-] | 455. | 28.0 | 427. | 30.1 | 0.438 | 0.650 | 0.553 | 0.602 | |||
| MIROC-ESM | [-] | 349. | 23.5 | 323. | 24.8 | 5.32 | -0.156 | 0.568 | 0.598 | 0.583 | ||
| MIROC-ESM-CHEM | [-] | 351. | 23.9 | 323. | 24.8 | 5.32 | -0.103 | 0.574 | 0.570 | 0.572 | ||
| MPI-ESM-LR | [-] | 335. | 10.8 | 322. | 29.0 | 1.09 | -2.05 | 0.497 | 0.176 | 0.336 | ||
| MRI-ESM1 | [-] | 588. | 57.3 | 529. | 30.1 | 3.49 | 0.581 | 0.824 | 0.703 | |||
| NorESM1-M | [-] | 537. | 23.7 | 507. | 30.1 | -0.449 | 0.599 | 0.725 | 0.662 | |||
| NorESM1-ME | [-] | 548. | 23.7 | 517. | 30.1 | -0.448 | 0.596 | 0.731 | 0.663 |
Temporally integrated period mean