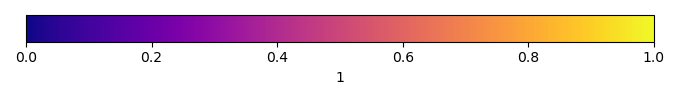Mean State
Download Data |
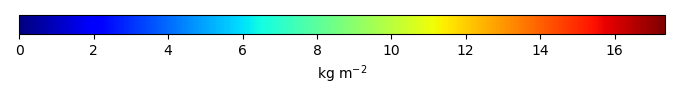
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (complement) [Pg] |
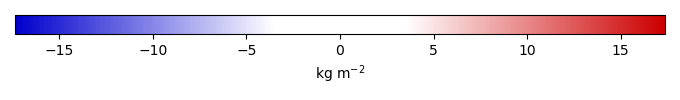
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 30.1 | ||||||||||
| bcc-csm1-1 | [-] | 476. | 25.2 | 30.1 | 443. | 0.000275 | -0.292 | 0.376 | 0.368 | 0.372 | ||
| BCC-CSM2-MR | [-] | 386. | 22.0 | 29.9 | 363. | 0.201 | -0.738 | 0.500 | 0.695 | 0.597 | ||
| CanESM2 | [-] | 529. | 21.3 | 30.1 | 495. | -0.948 | 0.513 | 0.730 | 0.622 | |||
| CanESM5 | [-] | 509. | 33.0 | 30.1 | 467. | 0.805 | 0.399 | 0.849 | 0.624 | |||
| CESM1-BGC | [-] | 529. | 18.8 | 29.9 | 509. | 0.176 | -1.03 | 0.552 | 0.659 | 0.605 | ||
| CESM2 | [-] | 485. | 16.4 | 29.9 | 467. | 0.176 | -1.18 | 0.491 | 0.678 | 0.585 | ||
| GFDL-ESM2G | [-] | 655. | 28.9 | 30.1 | 618. | 0.191 | 0.325 | 0.620 | 0.472 | |||
| GFDL-ESM4 | [-] | 405. | 20.5 | 30.1 | 385. | -1.05 | 0.403 | 0.507 | 0.455 | |||
| IPSL-CM5A-LR | [-] | 622. | 44.3 | 30.1 | 567. | 0.0202 | 2.24 | 0.341 | 0.780 | 0.560 | ||
| IPSL-CM6A-LR | [-] | 350. | 17.9 | 29.8 | 330. | 0.329 | -1.12 | 0.461 | 0.374 | 0.417 | ||
| MeanCMIP5 | [-] | 605. | 27.1 | 30.1 | 578. | -0.327 | 0.418 | 0.477 | 0.448 | |||
| MeanCMIP6 | [-] | 460. | 23.7 | 30.1 | 436. | -0.344 | 0.457 | 0.581 | 0.519 | |||
| MIROC-ESM | [-] | 349. | 23.5 | 24.8 | 323. | 5.32 | -0.156 | 0.408 | 0.598 | 0.503 | ||
| MIROC-ESM2L | [-] | 520. | 35.2 | 30.1 | 474. | 0.000179 | 0.975 | 0.331 | 0.830 | 0.580 | ||
| MPI-ESM-LR | [-] | 335. | 10.8 | 29.0 | 322. | 1.09 | -2.05 | 0.496 | 0.176 | 0.336 | ||
| NorESM1-ME | [-] | 548. | 23.7 | 30.1 | 517. | -0.448 | 0.487 | 0.731 | 0.609 | |||
| NorESM2-LM | [-] | 463. | 15.0 | 30.1 | 442. | -1.16 | 0.498 | 0.658 | 0.578 | |||
| UK-HadGEM2-ES | [-] | 453. | 19.1 | 29.8 | 431. | 0.282 | -0.818 | 0.525 | 0.664 | 0.595 | ||
| UKESM1-0-LL | [-] | 545. | 26.5 | 29.8 | 516. | 0.329 | 0.120 | 0.439 | 0.711 | 0.575 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 2.77 | ||||||||||
| bcc-csm1-1 | [-] | 66.5 | 1.58 | 2.76 | 65.9 | 0.00261 | -0.579 | 0.634 | 0.180 | 0.407 | ||
| BCC-CSM2-MR | [-] | 81.3 | 2.16 | 2.71 | 82.2 | 0.0568 | -0.228 | 0.619 | 0.728 | 0.673 | ||
| CanESM2 | [-] | 114. | 3.01 | 2.77 | 112. | 0.138 | 0.581 | 0.664 | 0.623 | |||
| CanESM5 | [-] | 87.0 | 0.664 | 2.77 | 87.6 | -1.07 | 0.637 | 0.651 | 0.644 | |||
| CESM1-BGC | [-] | 31.6 | 0.345 | 2.66 | 31.1 | 0.107 | -1.61 | 0.608 | 0.0337 | 0.321 | ||
| CESM2 | [-] | 111. | 5.20 | 2.66 | 105. | 0.107 | 2.43 | 0.296 | 0.842 | 0.569 | ||
| GFDL-ESM2G | [-] | 181. | 9.27 | 2.77 | 175. | 5.33 | 0.105 | 0.718 | 0.412 | |||
| GFDL-ESM4 | [-] | 162. | 8.52 | 2.77 | 153. | 3.93 | 0.104 | 0.532 | 0.318 | |||
| IPSL-CM5A-LR | [-] | 216. | 6.50 | 2.76 | 201. | 0.0109 | 3.57 | 0.359 | 0.521 | 0.440 | ||
| IPSL-CM6A-LR | [-] | 34.4 | 1.75 | 2.66 | 32.8 | 0.112 | -0.397 | 0.576 | 0.458 | 0.517 | ||
| MeanCMIP5 | [-] | 105. | 3.84 | 2.77 | 102. | 0.737 | 0.411 | 0.305 | 0.358 | |||
| MeanCMIP6 | [-] | 81.6 | 2.91 | 2.77 | 78.7 | 0.514 | 0.538 | 0.545 | 0.541 | |||
| MIROC-ESM | [-] | 80.4 | 3.97 | 1.72 | 77.3 | 1.05 | 2.05 | 0.241 | 0.701 | 0.471 | ||
| MIROC-ESM2L | [-] | 90.2 | 2.59 | 2.77 | 88.9 | 0.0138 | 0.573 | 0.778 | 0.675 | |||
| MPI-ESM-LR | [-] | 42.7 | 0.484 | 2.52 | 43.2 | 0.243 | -1.58 | 0.641 | 0.0463 | 0.344 | ||
| NorESM1-ME | [-] | 35.9 | 0.223 | 2.77 | 33.3 | -1.64 | 0.597 | 0.0304 | 0.313 | |||
| NorESM2-LM | [-] | 98.4 | 4.20 | 2.77 | 89.9 | 1.82 | 0.367 | 0.815 | 0.591 | |||
| UK-HadGEM2-ES | [-] | 50.8 | 2.43 | 2.54 | 50.4 | 0.230 | 0.712 | 0.459 | 0.750 | 0.605 | ||
| UKESM1-0-LL | [-] | 71.0 | 3.73 | 2.65 | 67.3 | 0.120 | 1.57 | 0.391 | 0.701 | 0.546 |
Temporally integrated period mean